A critical but constrained tool
Biomarker testing is central to oncology research and clinical care, driving targeted therapy development and personalized treatment. Next-generation sequencing (NGS) based approaches can detect diverse genomic alterations in a single assay, however, their broad adoption is hindered by the quality and quantity of available tumor samples.
The sample quality gap
A persistent challenge in biomarker testing is the reliance on formalin-fixed paraffin-embedded (FFPE) tissue samples. FFPE is the standard for specimen preservation, but extracting high-quality DNA and RNA for NGS is difficult.1-3 Many clinical samples—especially core needle biopsies—may yield only a few nanograms of usable material.
Dr. Erin Newburn, PhD, Director of Field Applications at Labcorp, highlights the widespread nature of this issue: “Clinical trial samples from FFPE blocks often have low nucleic acid yields and degradation. This impacts extraction, library preparation and post-sequencing data processing.”
Unlocking value with optimized workflows and scalable technologies
To address these limitations, labs are adopting standardized workflows that maximize data output from challenging samples. Labcorp has developed automated dual extraction protocols to improve nucleic acid yield and quality from FFPE tissues, reducing the number of quantity not sufficient (QNS) samples.4
“Automated, standardized workflows significantly improve nucleic acid yield and quality,” says Dr. Newburn. “This enables more successful genomic profiling and ultimately better patient outcomes.”
Beyond extraction, quality control (QC) metrics help ensure high-confidence variant calls.5 QC tools filter low-confidence data and minimize false results, providing reliable data to drive informed decisions.
Broad genomic solutions for maximum insight
Technologies including whole-exome sequencing (WES), whole-transcriptome sequencing (WTS) and large targeted panels are a key strategy for maximizing variant detection from minimal input.
Adds Dr. Newburn, "Comprehensive platforms help biopharma teams maximize data outputs for current and future biomarkers of interest from limited material.”
Best practices for success
To optimize comprehensive genomic profiling (CGP) in sample-constrained settings, early alignment with experienced genomics providers is key. Best practices include:
- Standardized extraction for reproducibility and scalability
- QC-informed library preparation to improve data quality and reduce rework
- Advanced sequencing platforms that deliver insights from low-input samples
These approaches improve outcomes, accelerate timelines and reduce costs—critical in clinical and research environments.
Case studies: Enabling high-confidence variant detection for your biomarker-driven trial
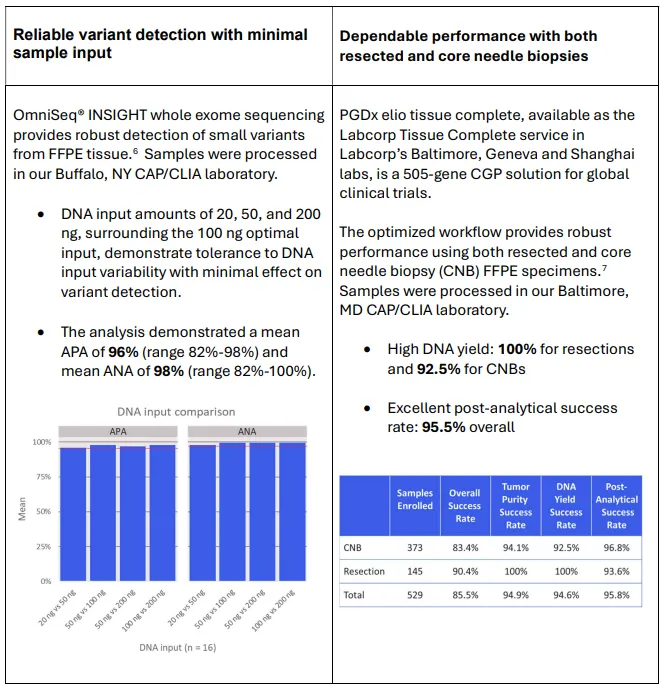
As CGP becomes integral to oncology development, generating reliable data from limited samples is essential. Through innovations in extraction, sequencing and analysis, Labcorp is helping unlock the full potential of precision oncology, one sample at a time.
Partner with Labcorp to accelerate your oncology trials with scalable, precision-driven solutions.
References
1 Hedegaard J, Thorsen K, Lund MK, et al. PLoS One. 2014;9(5):e98187. doi:10.1371/journal.pone.0098187
2 Spencer DH, Sehn JK, Abel HJ, Watson MA, Pfeifer JD, Duncavage EJ. J Mol Diagn. 2013;15(5):623-633. doi:10.1016/j.jmoldx.2013.05.004
3 McDonough SJ, Bhagwate A, Sun Z, et al. PLoS One. 2019;14(4):e0211400. doi:10.1371/journal.pone.0211400
4 Amirault K, Collins M, Beker L, et al. SLAS Technol. 2025;31:100252. doi:10.1016/j.slast.2025.100252
5 Do H, Dobrovic A. Clin Chem. 2015;61(1):64-71. doi:10.1373/clinchem.2014.223040
6 An J, Collins MJ, DeBlasi EA, et al. Published 23 November 2024 https://oncology.labcorp.com/analytical-validation-omniseqr-insight-whole-exome-sequencing-assay-facilitate-precision-oncology
7 Data on file, Labcorp